Revolutionizing MicroRNA Detection with One-step Method
MicroRNAs (miRNAs) are a class of small RNA molecules encoded by endogenous genes, typically 19–25 nucleotides (nt) in length. These non-coding RNAs, despite not encoding proteins, play crucial roles in regulating post-transcriptional gene expression. They are integral to processes such as cell proliferation, apoptosis, lipid metabolism, and differentiation. Dysregulation of miRNAs is closely linked to various diseases, particularly cancer, making them highly valuable as non-invasive biomarkers for clinical diagnostics.
Traditional MicroRNA Detection Methods
Several methods are employed for miRNA detection, each with its strengths and limitations:
• Northern blotting: A traditional technique, but complex, labor-intensive, prone to cross-contamination, and offers low sensitivity.
• Microarrays and RNA sequencing: Allow multiplex detection but are limited by their linear detection range and high costs.
• Sensor detection systems: Enable multiplex miRNA detection but often lack sufficient sensitivity and specificity.
• RT-qPCR: Including popular methods such as stem-loop RT-qPCR and poly(A)-tailed RT-qPCR, is widely recognized as the gold standard for miRNA detection. However, there are still limitations such as operational complexity and non-specific amplification.
Introducing SMOS-qPCR: An Innovative MicroRNA Detection Method
SMOS-qPCR (Sensitive and Multiplexed One-Step RT-qPCR) offers a groundbreaking solution for miRNA detection by combining multiplexed reverse transcription and TaqMan-based qPCR detection in a single reaction tube. This streamlined approach minimizes the risk of contamination from pipetting and simplifies the workflow.
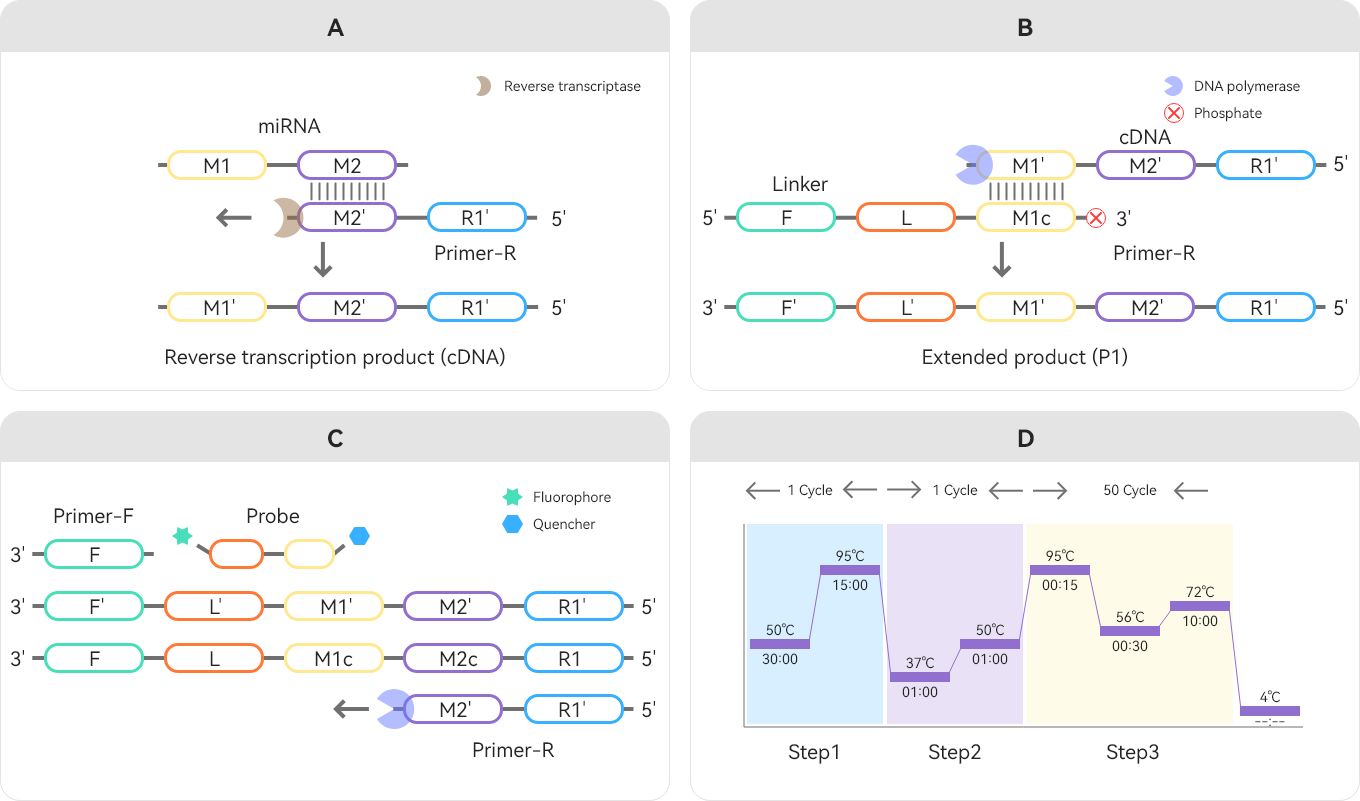
Fig. 1. The principle and work flowchart of the SMOS-qPCR system
The SMOS-qPCR method employs just two primers (Primer-F and Primer-R), a linker to extend the target template sequence, and a TaqMan probe. Through three key steps—reverse transcription, template extension, and qPCR amplification—it enables one-step detection of the target miRNA.
SMOS-qPCR Method and Fapon RT-qPCR Master Mix: The Optimal Companion for MicroRNA Detection
SMOS-qPCR utilizes Fapon RT-qPCR Master Mix, integrates multiplexed reverse transcription and TaqMan-based qPCR into a single tube, employing a one-step operation on a real-time PCR system.
Key Features of this system:
(1) High Sensitivity: Detects even low-abundance miRNAs with inputs as low as 28 femtograms (fg).
(2) Rapid Reaction: Completes the entire process in approximately 90 minutes, reducing hands-on time and contamination risk.
(3) Multiplex Capability: Analyzes up to four miRNAs in a single reaction, improving experimental efficiency.
(4) Robust Stability: Delivers consistent and reproducible results for reliable data interpretation.
Fapon's RT-qPCR Master Mix enhances miRNA detection within the SMOS-qPCR system and extends its utility to other RNA types, such as viral RNA and mRNA. Its superior performance, cost-effectiveness, and broad application make it an indispensable tool for RNA research and clinical diagnostics.
Driving Innovation in MicroRNA Research
Fapon is dedicated to pushing the boundaries of miRNA research and diagnostics through continuous technological advancements. With the innovative SMOS-qPCR system and our high-performance RT-qPCR reagents, we aim to empower researchers and clinicians in their pursuit of groundbreaking discoveries.
For inquiries or collaboration opportunities, please contact us at market@fapon.com. We look forward to partnering with you to advance the field of miRNA detection and clinical diagnostics.